Abstract
Background: Acute Myeloid Leukemia (AML) represents a group of heterogeneous clonal disorders of hematopoietic progenitor cells that are driven by multiple genetic events that impair cell differentiation and apoptosis and invoke uncontrolled cell proliferation. Consequently, immature myeloid blasts rapidly accumulate in the bone marrow and block the generation of mature end stage cells, which eventually causes potentially fatal infections and hemorrhages. AML is characterized by extensive genetic and phenotypic heterogeneity, which currently is diagnostically classified based on blast morphology defined by the French-American-British (FAB) Cooperative Group, cytogenetic abnormalities, and improved knowledge of genomic and epigenomic alterations. However, this has not yet resulted in treatment progress. AML patients are still routinely treated with a standard first-line combinatorial chemotherapy regimen with an overall survival rate remaining below 20%. Therefore, there is an unmet need for novel methods to stratify patients into therapy-relevant subtypes.
Concept and Aims: We hypothesize that phenotypic heterogeneity of AML is a consequence of dysregulated differentiation and that classification based on transcriptional reminiscence of cell of origin (COO) during AML transformation is of pathogenetic impact and therefore provide diagnostic and prognostic value. It is the aim of this project to generate and validate a COO classification system based on normal myeloid-cell-subset-associated gene signatures (MAGS) to stratify AML patients into therapy-relevant and prognostic subtypes.
MAGS Generation: Based on gene expression profiles of publicly available sorted normal myeloid subpopulations, we generated a COO classifier including the CD34+/CD38- hematopoietic stem cells (HSC), CD34+/CD38+/CD45RA- megakaryocyte-erythroid progenitors (MEP), and CD34+/CD38+/CD45RA+ granulocytic-monocytic progenitors (GMP) using regularized multinomial regression with 3 discrete outcomes and the elastic net penalty. The regularization parameters were chosen by cross validation.
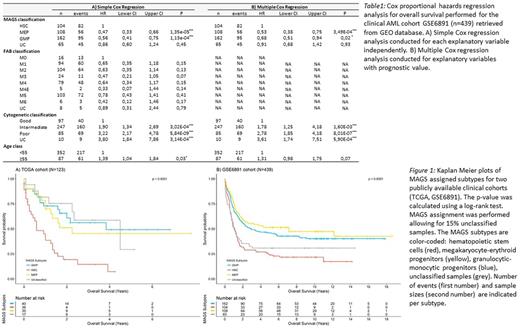
Results: MAGS prediction accuracy was validated in an independent dataset (n = 38; accuracy = 0.79) of sorted normal myeloid subpopulations. The prognostic value of MAGS assignment was studied by survival analyses using the Kaplan Meier method, log-rank test statistics, and simple and multiple Cox proportional hazards regression analysis in 2 clinical cohorts (TCGA: n = 115; GSE6891: n = 431). MAGS assignment had significant prognostic associations with overall survival (Figure 1; Table 1A) revealing that the linage committed MAGS subtypes GMP and MEP showed superior prognosis compared to the undifferentiated AMLs captured by the MAGS HSC subtype. Furthermore, multiple Cox regression analysis with MAGS assignment (HSC, GMP, MEP), FAB score (M0-M6), cytogenetics (good, intermediate, poor), and age (dichotomized into <55 and ≥55) as independent explanatory variables showed that MAGS assignment provides independent prognostic information (Table 1B), indicating that the MAGS classification captures distinct pathogenetic and prognostic knowledge not already explained by the FAB- or cytogenetic classifications. Molecular characterization of MAGS subtypes by differential gene expression analysis, gene set enrichment analysis, and identification of subtype specific mutation patterns of well-documented AML driver mutations indicated a reduced proliferation rate as well as overrepresentation of EVI1 mutations associated with the poor responding HSC subtype.
Conclusion: In accordance with our previous reports in malignant B-cell disorders, we have documented a novel classification system that associates normal myeloid progenitor subsets with AML subtypes of distinct pathogenetic and prognostic importance not already explained by the FAB- or cytogenetic classifications. Further pathogenetic studies are needed to develop a diagnostic platform using MAGS-assigned subtypes to improve disease management.
Severinsen: ARIAD: Research Funding; Celgene: Research Funding; Novartis: Research Funding. El-Galaly: Roche: Other: Travel funding; Takeda: Other: Travel funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal